Biomedical image analysis
The success of deep learning in image analysis has encouraged the biomedical imaging researchers to investigate its potential in analyzing various medical modalities to aid clinicians in faster diagnosis and treatment of diseases or infections like the on-going pandemic of SARS-CoV-2 (COVID-19). Following the deep learning usecases, the implication of classification can ascertain the presence or absence of disease in some modality e.g. the ground glass opacification (GGO) in the lungs via CT imaging. Furthermore, in localization, normal anatomy can be identified e.g. lungs in the CT or X-ray imaging, and later segmentation can generate refined boundaries around the GGOs to understand its impact on the anatomical structures for further analysis. Since, segmentation is an extension to classification, localization or detection, it offers very rich information about the disease and infected regions. With this interest, many architectures have been proposed for the segmentation of the targeted regions from vivid modalities. In addition, segmentation is the most widely researched application of deep learning in biomedical image analysis, where U-Net based segmentation architectures have gained significant popularity to develop computer-aided diagnosis (CAD) systems.
More details can be found in the following paper:
Punn, Narinder Singh and Sonali Agarwal. Modality specific U-Net variants for biomedical image segmentation: a survey. Artificial Intelligence Reviews (2022). https://doi.org/10.1007/s10462-022-10152-1
Summary of popular datasets for biomedical image segmentation
| Dataset | Description | URL |
|---|---|---|
| ISBI 2012 | Electron microscopy cell slides for cell segmentation | http://brainiac2.mit.edu/isbi_challenge/ |
| ISBI | 2D and 3D videos of moving cells for cell tracking | http://celltrackingchallenge.net/ |
| KDSB 2018 | Histopathological cell images for nuclei | https://www.kaggle.com/c/data-science-bowl-2018 |
| PanNuke | Histopathological slides for nuclei segmentation | https://jgamper.github.io/PanNukeDataset/ |
| DRIVE | Retinal fundus images for vessel | https://drive.grand-challenge.org/ |
| STARE | Retinal fundus imaging for blood vessel | http://cecas.clemson.edu/%7Eahoover/stare/ |
| CHASE_DB1 | Retinal fundus imaging for blood vessel | https://blogs.kingston.ac.uk/retinal/chasedb1/ |
| LiTS | Liver CT scans for tumor segmentation | https://competitions.codalab.org/competitions/17094 |
| LIDC-IDRI | Lung CT scans for cancer segmentation | https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI |
| LUNA 2016 | CT scans for lung nodule segmentation | https://luna16.grand-challenge.org/ |
| xVertSeg | CT spine images for vertebra segmentation | http://lit.fe.uni-lj.si/xVertSeg/ |
| SIIM-ACR | Chest X-rays for pneumothorax segmentation | https://www.kaggle.com/c/siim-acr-pneumothorax-segmentation/data |
| ISIC | Dermoscopy images for skin lesion segmentation | https://www.isic-archive.com/ |
| BraTS 2012 – 2020 | MRI modalities (T1, T2, FLAIR) for brain tumor segmentation | http://braintumorsegmentation.org/ |
| ISLES | MRI scans for stroke lesion segmentation | http://www.isles-challenge.org/ |
| ICCVB | Prostate MRI and retinal fundus imaging | http://i2cvb.github.io/ |
| IBSR | Repository of MRI imaging | https://www.nitrc.org/projects/ibsr |
| ACDC 2017 | MRI imaging for cardiac diagnosis and segmentation | https://www.creatis.insa-lyon.fr/Challenge/acdc/index.html |
| PROMIS 2012 | Prostate MRI image segmentation | https://promise12.grand-challenge.org/ |
| Medical Segmentation Decathlon | MRI and CT modalities for tumor segmentation in various organs like liver, brain, lung, etc. | http://medicaldecathlon.com/ |
| OASIS | MRI and PET images for aging analysis and segmentation | https://www.oasis-brains.org/ |
| Head-Neck-PET-CT | PET and CT imaging for tumor segmentation | https://wiki.cancerimagingarchive.net/display/Public/Head-Neck-PET-CT |
| BUSIS | Ultrasound imaging for breast tumor segmentation | http://cvprip.cs.usu.edu/busbench/ |
| BUSI | Breast ultrasound scans for tumor segmentation | https://scholar.cu.edu.eg/?q=afahmy/pages/dataset |
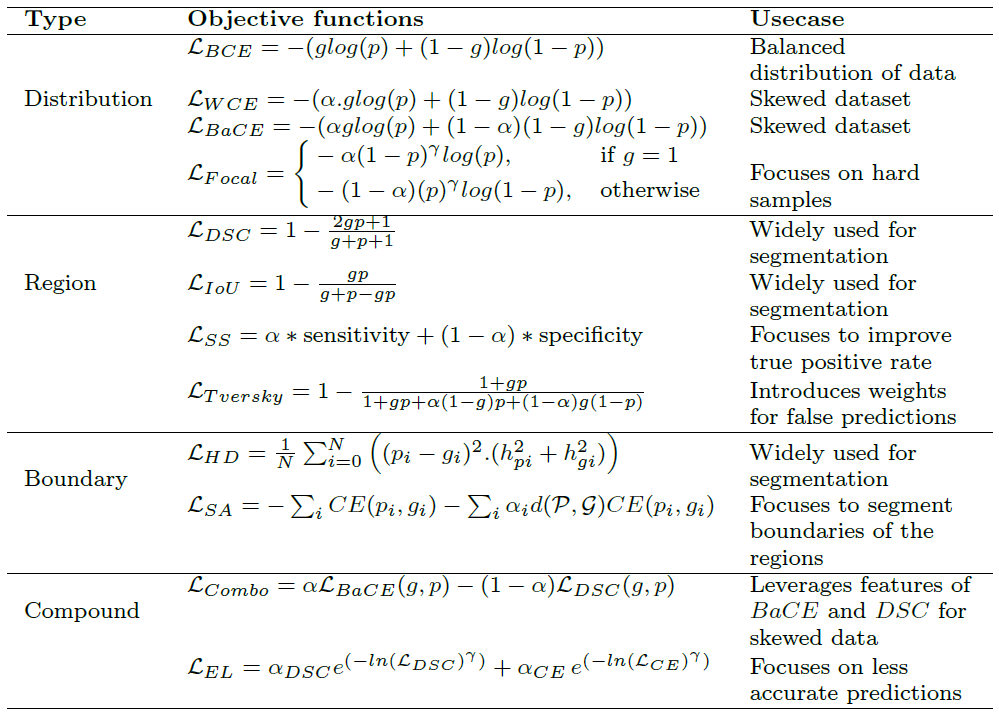
Summary of performance metrics for biomedical image segmentation
Summary of performance metrics for BIS in terms of number of true positive (![]() ), true negative (
), true negative (![]() ), false positive (
), false positive (![]() ) and false negative (
) and false negative (![]() ), predicted mask (
), predicted mask (![]() ) and ground truth (
) and ground truth (![]() ),
), ![]() is the directed
is the directed ![]() from
from ![]() to
to ![]() with
with ![]() as euclidean distance,
as euclidean distance, ![]() and
and ![]() refer to the volumes of generated and reference segmentation.
refer to the volumes of generated and reference segmentation.
In BIS, mostly the datasets are imbalanced i.e. the number of pixels/voxels concerning the target region (region of interest) are relatively less than the dark pixels/voxels (background region), due to which the metrics such as accuracy, which are best suited for a balanced distribution of data samples, are not recommended for BIS evaluation of the models. Among the shown metrics intersection-over-union (IoU or Jaccard index) and dice similarity coefficient are the most widely used evaluation metrics in BIS for various modalities. More details can be found here.
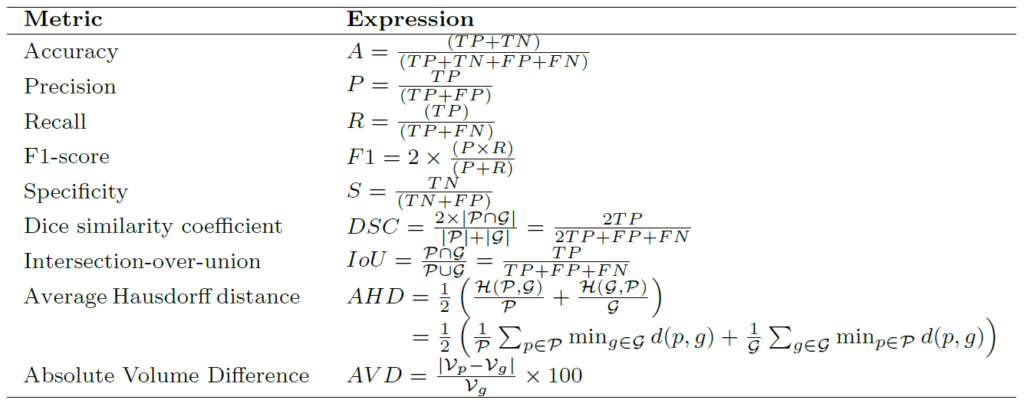
Summary of popular loss functions for biomedical image segmentation
Below table shows the summary of loss functions for biomedical image segmentation with respect to the predicted mask (![]() ) and ground truth mask (
) and ground truth mask (![]() ),
), ![]() and
and ![]() as constants,
as constants, ![]() is Hausdorff distance and
is Hausdorff distance and ![]() is the operator for Euclidean distance. More details can be found here.
is the operator for Euclidean distance. More details can be found here.