Publication Details
The paper is published in the journal of ACM Transactions on Multimedia Computing, Communications, and Applications (TOMM) in 2020: click here
Abstract
Most of the analysis requires nuclei detection as the starting phase for getting an insight into the underlying biological process and further diagnosis of diseases. Following this a novel architecture is proposed to generate segmentation masks of neuclie present in the histopathological images.
Highlights
- An Inception U-Net (IU-Net) model is proposed for biomedical image segmentation.
- The proposed architecture consists of switch normalization layer, convolution layers, hybrid pooling layers and inception layers (concatenated 1×1, 3×3, 5×5 convolution and the hybrid of max and Hartley spectral pooling layer).
- The model is trained with segmentation loss function defined as a weighted average of binary cross entropy loss, dice coefficient loss and intersection-over-union loss.
- The model is evaluated on Kaggle Data Science Bowl Challenge 2018 (KDSB18).
- The source code is available on my github page here.
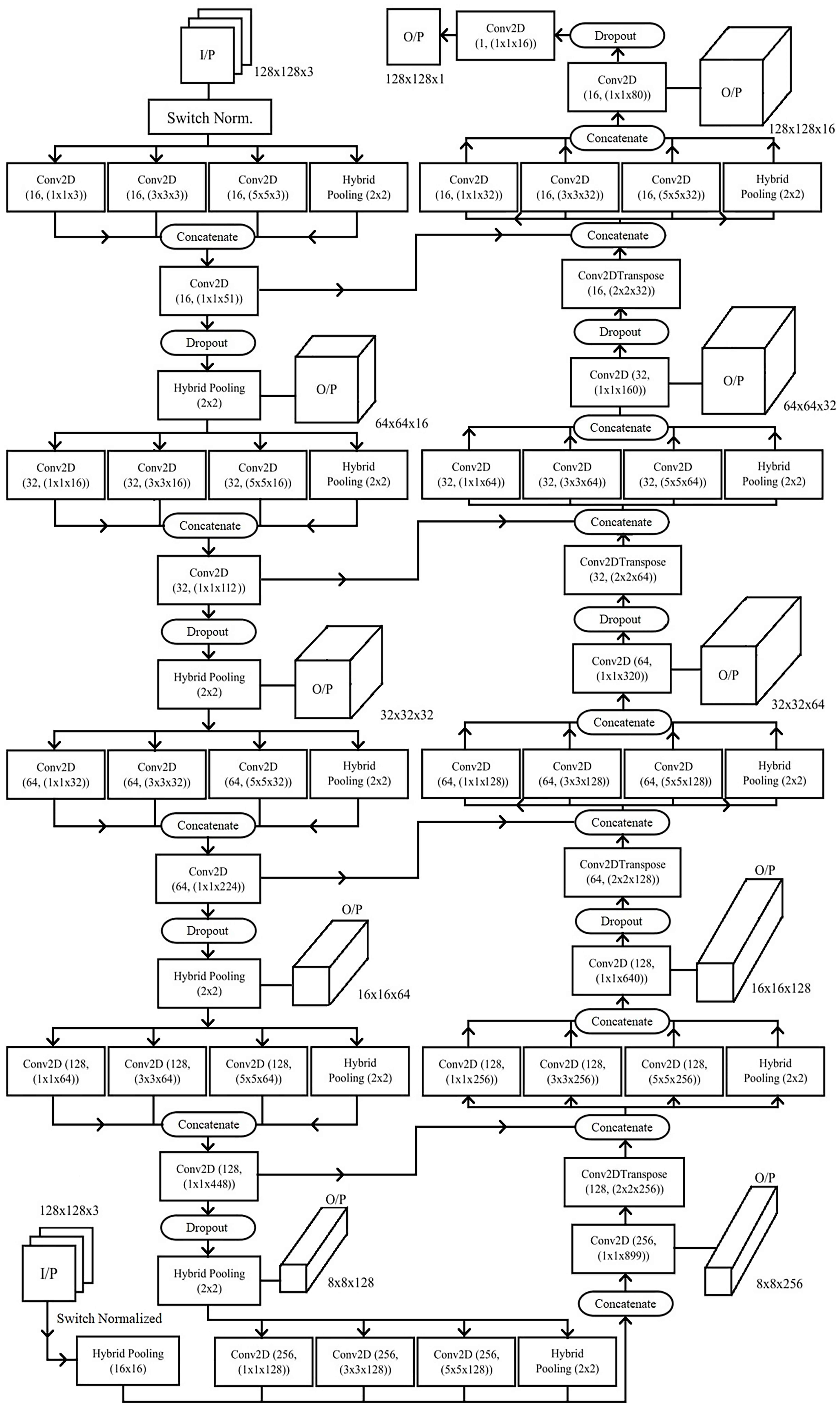
IU-Net Overview
The proposed inception U-Net architecture (as shown in Fig. 1) is the hybrid of excellent features of the two popular state-of-the-art architectures of deep learning, Google’s inception architecture and the U-Net architecture. The default convolution layers in U-Net architecture are replaced with the inception layers of GoogLe-Net, and also the max-pooling layers are replaced with the novel hybrid of Hartley spectral pooling and max pooling layer. For the training phase, the input is normalized with switch normalization that automatically adapts different normalization operations, i.e., batch norm, instance norm and layer norm with end-to-end learning. The segmentation loss is optimized as the weighted average of binary crossentropy, dice coefficient and mean intersection over union (IoU) which is later evaluated with pixel accuracy, loss value and IoU. The IoU score calculation/implementation is as per the Kaggle Data Science Bowl Challenge 2018 (KDSB18), which is the more precise and accurate approach for computing IoU. The U-Net inception model is implemented in python with the Keras version 2.2.4 and the Tensorflow-gpu version 1.12.0.
Output
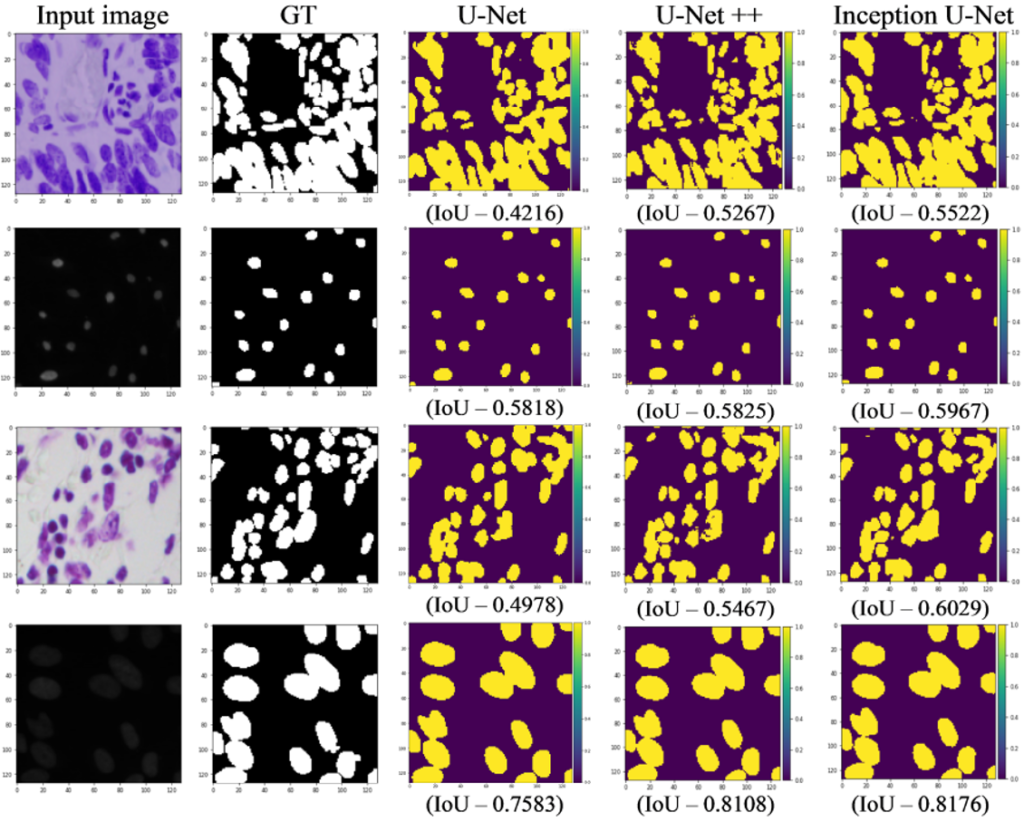
Below Fig. 2 shows the segmentation results on the KDSB18 challenge.
Findings
The inception U-Net architecture achieved good nuclei segmentation results on Kaggle Data Science Bowl 2018 challenge. With our proposed framework and segmentation loss function we achieved the IoU score of 65.23%. Experimentally it is shown that the patterns and the features extracted using blocks of inception and hybrid pooling layers are effective enough to improve upon the IoU score with the help of proposed segmentation loss function based on binary cross entropy, dice coefficient, and IoU score. Though this architecture is developed for nuclei segmentation, we believe that the same concept can further be extended to other biomedical imaging task and even in other deep learning areas.
For more details please refer my paper here.
