Publication details
The paper is published in the journal of Springer Applied Intelligence in 2020: click here.
Abstract
The novel coronavirus 2019 (COVID-19) is a respiratory syndrome that resembles pneumonia. Motivated from the highly contagious nature of COVID-19, we proposed a deep learning based computer-aided diagnosis system to classify COVID-19 infected samples using chest X-ray imaging.
Highlights
- With limited availability of the COVID-19 samples, we proposed random oversampling and weighted class loss function approach for unbiased fine-tuned learning (transfer learning). Furthermore, the data samples are mixed with the other randomly selected CXR images selected from another dataset, RSNA and NLM(MC).
- The proposed dataset is available on my GitHub page here.
- Various state-of-the-art deep learning approaches are considered such as ResNet, Inception-v3, Inception ResNet-v2, DenseNet169, and NASNetLarge.
- The problem is divided into binary classification (as normal and COVID-19 cases) and also multi-class classification (as COVID-19, pneumonia, and normal case) of posteroanterior CXR images.
- A new dataset is proposed that combines.
Overview
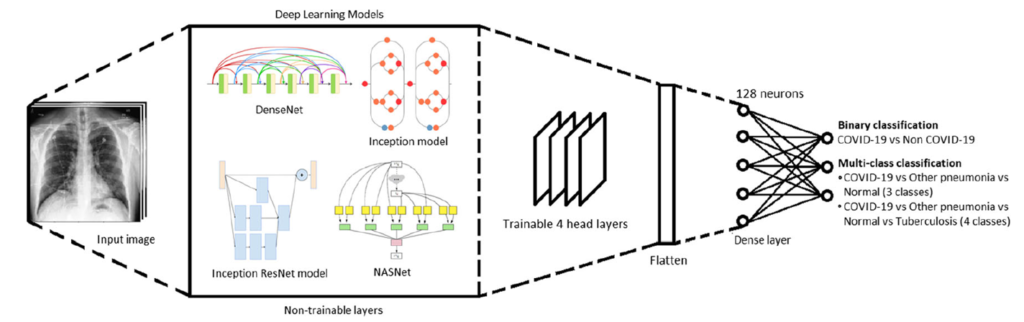
The overall proposed framework is shown in Fig. 1. Transfer learning is utilized on the state-of-the-art architectures to utilize the pre-trained models while fine tuning the head layers to serve the purpose of classifying the COVID-19 samples. This follows by enabling the four head layers of the network to adjust its trainable parameters along with the addition of fully connected layers with 128 neurons and 2 or 3 neurons in the output layer depending on binary or multi-class classification, accompanied with the softmax activation function. The classified results are then visualized via local interpretable model-agnostic explanations (LIME).
Output
Below Fig. 2 shows COVID-19 classification results with NASNetLarge model and LIME visualization.
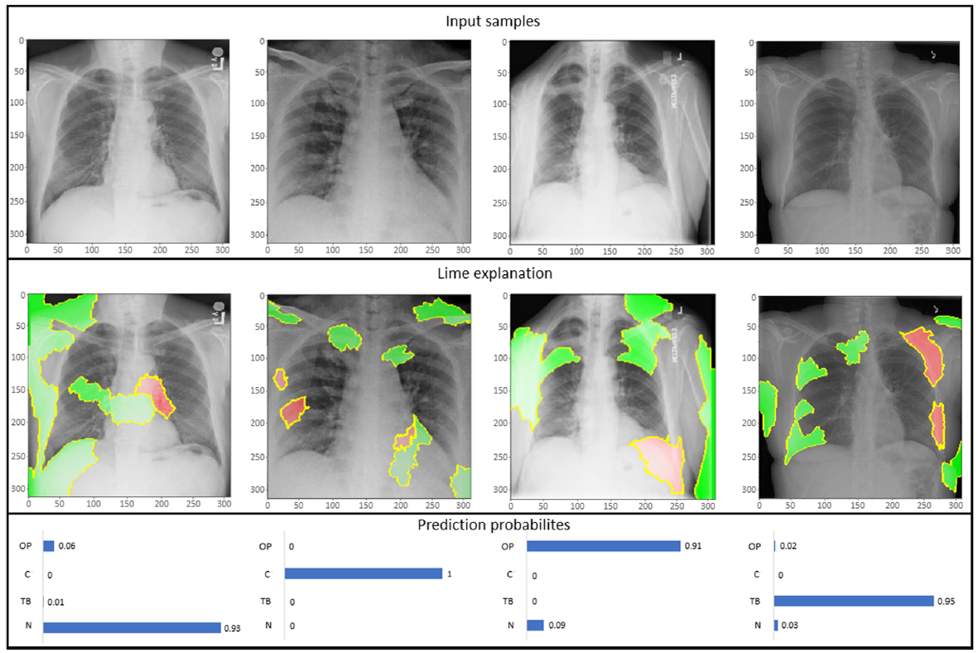
Findings
The models achieved different performance in different scenarios (binary/multi-class classification and oversampling/wegihted loss), among which NASNetLarge displayed better performance specially in binary classification of COVID-19 samples. The visual representation based on local interpretable model agnostic explanations is utilized to understand the basis of prediction of the model. As an extension to this work more deep learning models and preprocessing techniques can be explored to achieve better results.
For more details please refer my paper here.
