Publication details
The paper is published in the journal of Springer Multimedia Tools and Application in 2020: click here.
Abstract
For any radiologist, identification and segmentation of brain tumor (gliomas) via multi-sequence 3D volumetric MRI scan for diagnosis, monitoring, and treatment, are complex and time-consuming tasks. Hence an automated deep learning based approach is proposed to generate the segmentation mask to identify and localize the brain tumor using MRI.
Highlights
- The brain tumor segmentation is performed on the preprocessed multi-modalities by proposed 3D deep neural network framework.
- The framework comprises of several components such as multi-modalities fusion, tumor extractor, and tumor segmenter.
- The model is evaluated on BraTS 2017 and BraTS 2018 datsets.
- The source code is available on my GitHub page here.
Overview
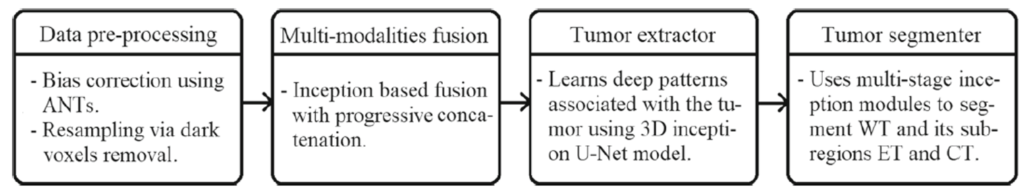
The components of the proposed framework are presented in Fig. 1. The overall architectural design consists of inception module, data preprocessing, multimodality encoded fusion, tumor extractor and tumor segmenter units with 10.5M trainable parameters. The preprocessed multi-modalities are progressively fused to gain multi-information that enables the tumor extractor component to analyse the deep patterns using the contraction and expansion pathways associated with the tumor regions. The tumor regions are segmented into whole tumor (WT), core tumor (CT), and enhancing tumor (ET) by tumor segmenter component that merges the features learned at different hierarchical layers of tumor extractor component with the help of upsampling operation.
Output
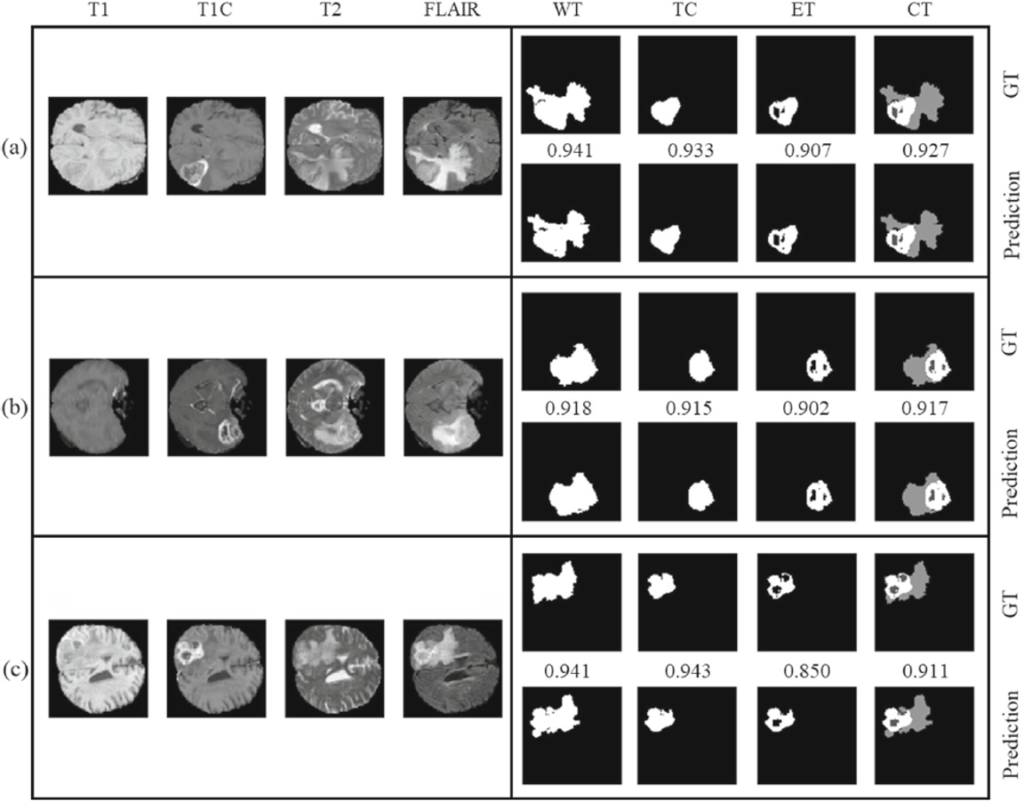
Below Fig. 2 shows the segmentation results on the BraTS Challenge.
Findings
The complex task of brain tumor segmentation is divided into multiple components: data-preprocessing, multi-modalities fusion, tumor extractor, and tumor segmenter. The proposed approach achieved significant improvements on BraTS 2017 and BraTS 2018 datasets by exploiting the advantages of inception convolutions, 3D U-Net architecture, and segmentation loss function that is based on Jaccard index and dice coefficient. As an extension, the work of the present article could further be improved by employing hybrid, cascaded, ensembled or other learning approaches.
For more details please refer my paper here.
