Publication details
The paper is published in the Special Issue of the ECML PKDD 2022 Journal Track – Springer Machine Learning as in 2022: click here
Abstract
In biomedical image segmentation, one of the major challenges is concerned with the limited annotated data availability. Motivated by the emerging self-supervised learning technology, we address this issue by proposing a BT-Unet framework that can be trained efficiently with limited samples for biomedical image segmentation. The framework exhibits the potential to enhance the segmentation performance of the U-Net models. The framework performs pre-training of U-Net encoder network with redundancy reduction based Barlow Twins strategy to learn feature representations in an unsupervised manner and later the U-Net model is fine-tuned for downstream segmentation task with limited annotated samples. This framework reduces the dependency of the U-Net models for a huge amount of training data to achieve the best segmentation results.
Highlights
- The challenge of limited biomedical annotated data availability is addressed by integrating redundancy reduction based self-supervised learning approach with U-Net segmentation models.
- The pre-training of the U-Net encoder is performed with the Barlow Twins strategy to learn feature representations in an unsupervised manner (without data annotations).
- The effect of pre-training on biomedical image segmentation performance is analyzed with multiple U-Net models over diverse datasets.
- The source code is available on my GitHub page here.
Overview
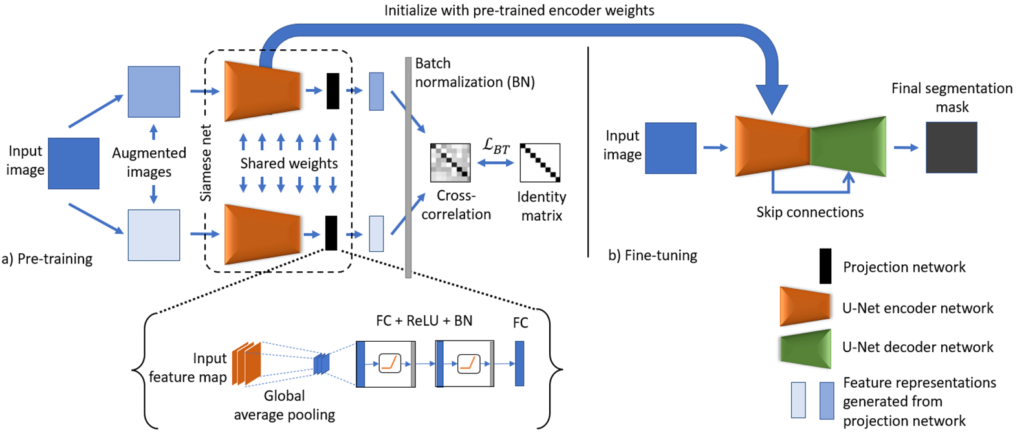
The schematic representation of the proposed model is shown in Fig. 1. The BT-Unet framework is divided into two phases: 1) Pre-training and 2) Fine-tuning. In pre-training, the aim is to learn the complex feature representations using unannotated data samples. Here, the encoder network of the U-Net model is pre-trained using BT self-supervised learning strategy. Initially, the input image is augmented or corrupted with certain distortions such as random crop and rotations to generate two distorted images. Each augmented image is analyzed with a U-Net encoder followed by a projection network to generate encoded feature representations in desired dimensions. The projection network follows from the feature maps produced by the encoder network with global average pooling and blocks of fully connected layers, ReLU activation and batch normalization (FC + ReLU + BN), and final encoded feature representations are generated by another FC layer. Following from the empirical observations, the number of neurons in each fully connected layer is kept half of the spatial dimension of an input image for efficient pre-training, e.g., if input, ![]() then the number of neurons are
then the number of neurons are ![]() , where
, where ![]() is a spatial dimension of an image. The number of neurons could be further increased but at the cost of heavy computation. However, no significant improvement was observed with increased dimensions. Since, in later layers, the network learns task specific features that are not aligned with the downstream segmentation task, hence the weights learned by the projection network can be neglected, whereas the weights of the entire encoder network can be transferred to the U-Net model. In the second phase, the weights of the encoder network in the U-Net model are initialized with pre-trained weights (from the first phase), whereas the rest of the network is initialized with default weights. Finally, the U-Net model is fine-tuned with limited annotated samples for the biomedical image segmentation.
is a spatial dimension of an image. The number of neurons could be further increased but at the cost of heavy computation. However, no significant improvement was observed with increased dimensions. Since, in later layers, the network learns task specific features that are not aligned with the downstream segmentation task, hence the weights learned by the projection network can be neglected, whereas the weights of the entire encoder network can be transferred to the U-Net model. In the second phase, the weights of the encoder network in the U-Net model are initialized with pre-trained weights (from the first phase), whereas the rest of the network is initialized with default weights. Finally, the U-Net model is fine-tuned with limited annotated samples for the biomedical image segmentation.
Output
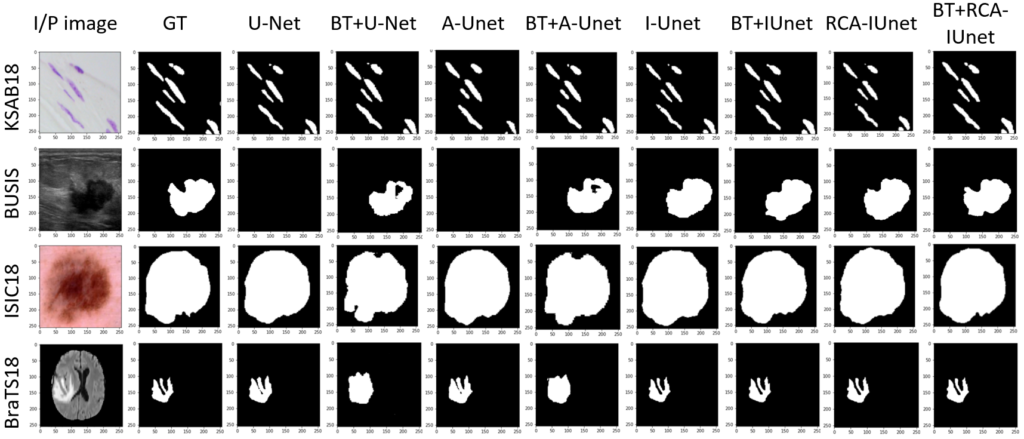
Below Fig. 3 shows the segmentation results of the different segmentation models with and without the pre-training over diverse datasets. The models that are considered for experiments are vanilla U-Net, attention U-Net (A-Unet), inception U-Net (I-Unet) and residual cross-spatial attention guided inception U-Net (RCA-IUnet).
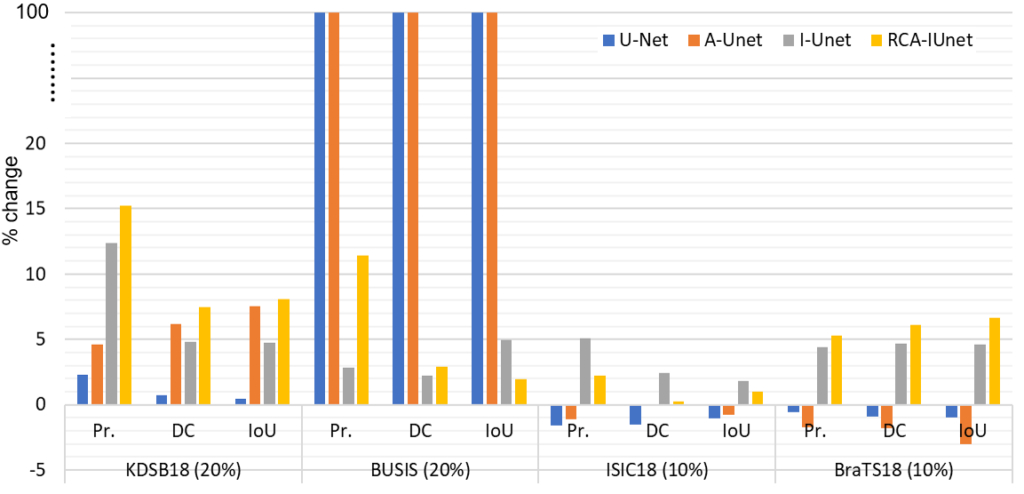
The framework is evaluated on four different biomedical imaging datasets and U-Net models, where we analyzed the percentage change in the performance of all the models with and without the Barlow Twins pre-training as shown in Fig. 3. For instance, the performance of the RCA-IUnet model increases in terms of dice coefficient (most common metric to measure segmentation performance) with 7.5%, 3.0%, 0.2% and 6.1% in KDSB18, BUSIS, ISIC18 and BraTS18 datasets respectively.
Findings
The BT-Unet framework uses redundancy reduction based Barlow Twins strategy for pre-training the encoder network of the U-Net model with feature representations of the data in an unsupervised manner, followed by fine-tuning of the U-Net model for downstream biomedical image segmentation task with limited annotated data samples. With exhaustive experimental trials, it is evident that BT-Unet tends to improve the segmentation performance of U-Net models in such situations. Moreover, this improvement is also influenced by the underlying encoder structure and nature of the biomedical image segmentation task.
